Circadian rhythm, observed in various metabolic and physiological processes, is governed by the endogenous clock found in the suprachiasmatic nucleus (SCN) of the hypothalamus and by peripheral clocks. The circadian system of the organism acts independently of external cues, but it can adapt to environmental conditions, such as light, temperature, meals/feeding etc. The circadian clock in the SCN is synchronized to external cues, and it coordinates the peripheral clock by sending signals to peripheral tissues (1). The molecular mechanism of the self-sustained circadian system of the organism involves circadian genes. Circadian genes presenting oscillatory transcriptional-translational feedback loops have been found not only in the SCN, but also in peripheral tissues, including human peripheral leukocytes. The biological clock is controlled by several genes, including BMAL1, CLOCK, Period (PER) and Cryptochrome (CRY) (2).
Some epidemiological studies have found that long-term, night-working women have a higher risk of breast cancer than women who do not work at night (3). One of several proposed mechanisms underlying cancer risk among individuals exposed to light at night involves circadian genes expression alteration (4). Circadian genes may play an important role in the control of multiple biological processes (5), including DNA repair, oxidative stress, maintenance of genomic stability, cell proliferation, and apoptosis (6, 7). Therefore, they may have important relevance to the carcinogenic process. Alteration of circadian genes was found to be associated with various disorders in humans, including sleep and mental disorders, cardiovascular diseases, obesity, impaired glucose tolerance, alcohol abuse, and cancer (8–12).
It has been widely observed that a molecular feedback loop exists for specific clock genes that act as positive and as well negative regulators of the biological clock. Moreover, circadian genes expression among humans is reported to be similar to that observed in rodent peripheral tissue, although tissue-specific clock genes expression patterns were also observed (1). The daily variations in expression of clock genes associated with different rhythmicity and maximal or minimal expression in various peripheral tissues of the human organism are the strongest for peripheral blood leukocytes, probably due to the presence of various cell populations in that tissue. However, blood leukocytes subpopulations may be useful for the investigation of human circadian rhythms as circadian genes are expressed with the peak level occurring during the habitual time of activity (13). In leukocytes, including peripheral blood mononuclear cells (PBMC), the acrophase of PER messenger ribonucleic acid (mRNA) was observed in the early morning and morning (13–19). CRY1 and CRY2 mRNA expression level peaked also in the morning in circulating leukocytes (20), while the positive regulator of circadian rhythm – BMAL1 mRNA – presented maximal expression in PBMC and leukocytes in the evening or midnight, as it is in approximate antiphase with the other negative regulators of circadian rhythm (15, 18, 20–22). Interestingly, in the majority of studies, various populations of blood leukocytes presented lack of rhythmicity of CLOCK mRNA expression (17, 20).
Humans are well synchronized to the external light–dark cycles. However, disturbances of these cycles by irregular sleep hours, shift work, or light at night exposure may induce alteration of the SCN and peripheral circadian rhythms. Therefore, the aim of our analysis, which forms part of a cross-sectional epidemiological study of nurses and midwives (23), was to determine the effect of rotating night shift work on the expression of selected core circadian genes: BMAL1, CLOCK (positive regulators), CRY1, CRY2, PER1, PER2, and PER3 (negative regulators) as indicators of peripheral clock.
Methods
Study population
A cross-sectional study of 354 nurses and midwives, all female, currently working rotating night shifts and 370 nurses who work only during the day was carried out in Lodz, Poland in 2008–2011 (23). Women were selected from the Local Registry of the Chamber of Nurses and Midwives. The questionnaire data and biological samples were collected in 2008–2010. The inclusion criteria for the nurses were age 40–60 years and current employment. Gene expression analysis was conducted among 92 pairs of nurses and midwives. The required number of rotating night shift workers were sampled from the study population. For each rotating night shift worker, one non-night shift nurse was randomly selected matched for age (within two years) and sampling season (March–September and October–February).
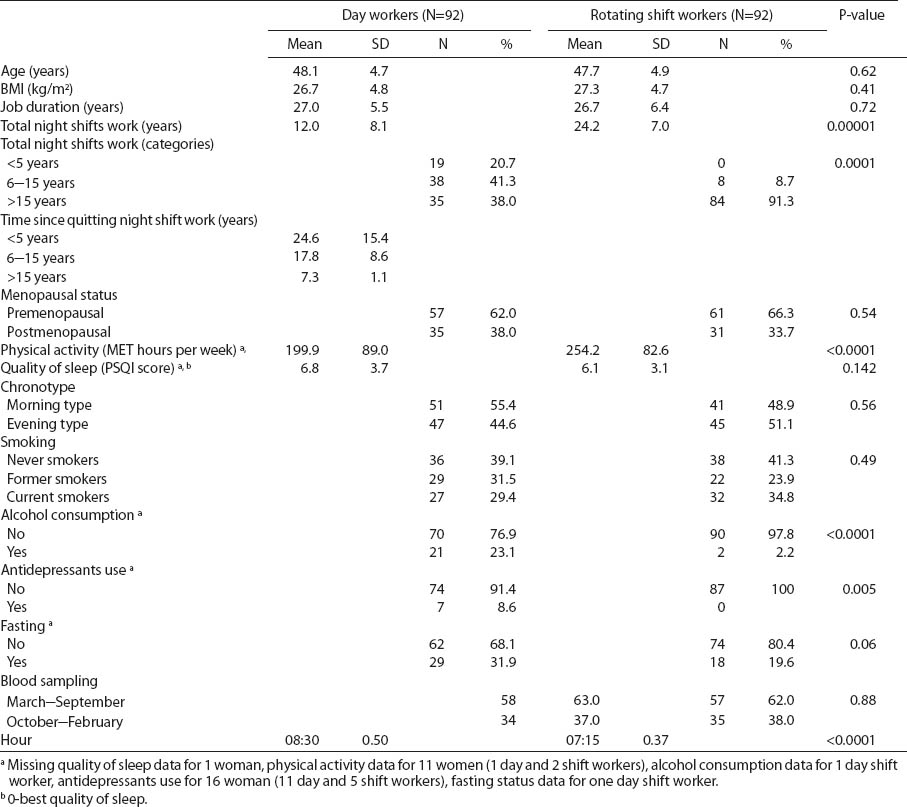
The study participants completed a structured questionnaire during in-person interviews. Data included information on demographics, occupational history, the current and previous jobs, physical activity [according to the International Questionnaire on Physical Activity (IPAQ)], sleep quality [according to the Pittsburgh Sleep Quality Index (PSQI)], chronotype (according to self-reported preference), tobacco smoking, and alcohol use. Characteristics for the two groups are shown in table 1.
Table 1
Selected characteristics of nurses and midwives under different work shifts. [BMI=body mass index; MET=metabolic equivalents of task; PSQI=Pittsburgh Sleep Quality Index; SD=standard deviation]
The Ethics Committee for Scientific Research at the Nofer Institute approved the study protocol, and a written informed consent was obtained from each study participant.
Gene expression analysis
Blood samples were collected into S-Monovette® heparinized test tubes (Sarstedt AG & Co, Nümbrecht, Germany) in the morning (06:00–10:00 hours). Transcript levels of BMAL1, CLOCK, CRY1, CRY2, PER1, PER2, and PER3 in peripheral blood leukocytes were determined by means of quantitative real-time polymerase chain reaction (PCR). Primers were designed with Beacon Designer 7.01 (PREMIER Biosoft International, Palo Alto, CA, USA). Amplicon sequence complied with exon-exon boundaries (table 2). Total RNA was isolated from whole blood, immediately after delivery to the lab, by means of QIAamp RNA Blood Mini Kit (Qiagen). cDNA was synthesized on 250 ng RNA with Quantitect Kit (Qiagen, Syngen Biotech, Hilden, Germany). Expression of circadian genes was quantified with FastStart SYBR Green Master (Roche, Basel, Switzerland) and using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the endogenous control, which presents stable expression levels in RNA isolated from peripheral blood leukocytes. Reference gene-normalized relative quantification with efficiency correction for target and reference gene (Pfaffl method) was calculated using Q-Gene, a Microsoft Excel®-based software application (Microsoft, Redmond, WA, USA) coded in Visual Basic (24). PCR efficiencies were calculated using dilutions of five pooled cDNA samples, which were reverse transcribed from total human RNA isolated from leukocytes. All the samples were amplified in duplicate. Randomly selected pairs of samples were simultaneously amplified on the same plate for the eight genes examined. To determine inter-assay variability, 12 samples (6 pairs) were run over 2 separate occasions for ≤30 days of the experiment. Inter- and intra-assays coefficients of variability (CV) for the examined eight genes (calculated using relative expression values) were <15% and <10%, respectively.
Statistical analysis
Arithmetic means with standard deviations (SD) and frequencies of the basic characteristics were calculated. The linear regression was used to compare means of the continuous variables, and the Pearson Chi-square test was calculated to compare the frequencies distribution among women working on rotating night shifts and day workers. The associations between rotating night shift work measures and circadian genes expression were computed by linear regression. All expression variables were log transformed except BMAL1 and CRY2. The list of covariates included age, current smoking status (yes/no), antidepressants use (yes/no), hour of blood collection, quality of sleep (expressed as PSQI), physical activity [expressed in metabolic equivalents of task (MET) hours per week], season of the year of blood collection (March–September/October–February), body mass index (BMI), menopausal status (self-reported pre-/postmenopausal), alcohol drinking the day before blood collection (yes/no), and fasting (<6 hours of blood collection/≥6 hours of blood collection). Of these covariates in the final models for each gene expression, only significant confounders were retained (the list of confounders is provided in tables 3 and 4). Stepwise selection method was used with the Akaike information criterion (AIC) to include variables. The value of P<0.05 for group characteristics was considered to represent statistical significance, and the statistical tests were two-sided. To account for multiple comparison (7 clock genes) we applied Bonferroni correction method with significance level of P<0.05/7=0.007.
Table 3
Circadian gene relative expression among nurses and midwives under different work shifts and correlation coefficients between gene expression and rotating night shift work.
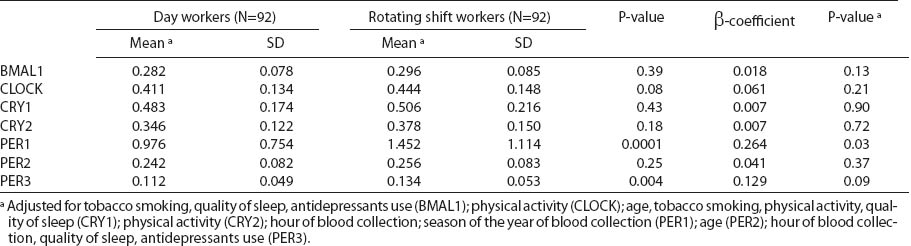
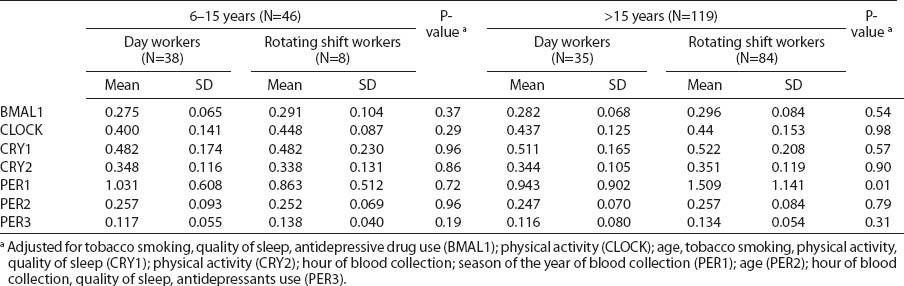
Table 4
Circadian gene relative expression and different categories of total night shift work in nurses and midwives under different work shifts. [SD=standard deviation]
Statistical analyses were performed with STATA11 (StataCorp LP, College Station, TX, USA) software.
Results
The average total shift work duration among women currently working rotating night shifts was significantly longer (24.2 years) than among current day workers (12.0 years). All of the women and midwives who were current day workers had previously worked rotating night shifts. The majority (91.3%) of current rotating night shift workers had been working night shifts for >15 years. The proportion of women currently working day shifts who had previously worked >15 years on a rotating night shift schedule was 38.0%, with an average of 7.3 years since quitting night shift work (table 1).
Rotating shift nurses and midwives drank alcohol beverages the day before collection significantly less frequently than day working women (P<0.0001). None of the rotating night shift nurses used antidepressants and women in this group were more physically active than day workers (P<0.0001).
Among rotating night shift women, blood samples were collected at the end of night shift at 07:15 (SD 0.37) hours and at the beginning of work [08.30 (SD 0.50) hours] for day working nurses (P<0.0001).
All subjects showed detectable transcript levels in blood leukocytes, however with large interindividual variation. For all analyzed circadian genes, the mRNA level of the particular gene presented positive correlation with the mRNA level of other genes in the whole study group and subgroups defined by rotating work status, age, BMI, menopausal status, smoking status, or chronotype.
PER1 and PER3 mRNA expression was associated with the hour of blood collection. Transcripts level was significantly down-regulated in the late versus early hours of the morning (06:00–10:00 hours) (β-coefficient -0.226, P=0.001 for PER1 and β-coefficient -0.181, P<0.0001 for PER3).
An elevated circadian gene expression was observed among rotating night shift compared with day workers. However, we did not observed statistically significant changes of investigated genes expression among nurses working night compared to day shifts, when confounding factors that significantly influenced particular gene expression were included in the analysis (table 3). In case of PER1, we observed a strong effect of collinearity with time of blood sampling, as both variables were of borderline significance but the joint test was very significant (P=0.00008).
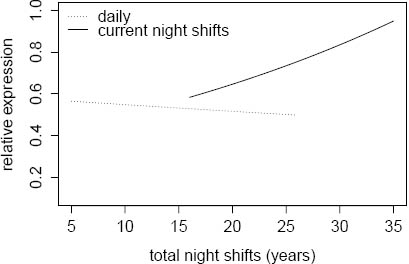
Circadian gene expression was not associated with the duration of rotating night shift work, specified as three categories of total night shift work (<5, 6–15, and >15 years) among women currently and previously (current day workers) working night shifts (table 4). The highest expression of PER1 mRNA was observed among current rotating shift women (N=84) who had worked a rotating night shift work schedule >15 years compared with day workers who had previously worked rotating shifts >15 years (N=35) (b-coefficient 0.264, P=0.03) (table 4). Moreover, the PER1 transcripts level was associated with the lifetime duration of rotating night shift work among current night shift workers (ß-coefficient 0.026, P=0.04), while we did not observe such an association among current day workers (ß-coefficient -0.006, P=0.50) (figure 1).
Discussion
Circadian gene expression patterns may be useful as peripheral clock biomarkers in circulating leukocytes. In the presented gene expression study, we analyzed circadian gene expression in peripheral blood leukocytes of nurses and midwives who worked irregular night shifts. To exclude the majority of factors and conditions that may possibly influence leukocyte gene expression (25), we analyzed randomly selected nurses matched for age (within 2 years) and sampling season (March–September and October–February). In the total study group and subgroups of current day workers and rotating night workers, significant correlations were observed between expression of positive and negative regulators of peripheral clock, regardless of menopausal status, smoking, alcohol use, chronotype, age, BMI, sleep quality or physical activity. It may confirm dependence of circadian genes in peripheral tissue as they are linked to the same process of transcriptional-translational feedback loop.
Further, we used a whole population of white blood cells, which includes mononuclear and polymorphonuclear cells, as it was found that there is no difference in the expression of PER1 mRNA between these two populations of white blood cells (16). However, it should be pointed out that different rhythmicity of clock gene expression was observed for peripheral blood leukocytes, probably due to the presence of various cell populations in that tissue (14–22). In the majority of in vivo studies, daily monitoring of the expression of clock genes in human peripheral leukocyte populations has been extensively investigated. In human peripheral leukocytes, clock genes have demonstrated expression rhythmicity with PER and CRY mRNA maximal expression in the early morning or morning and BMAL1 mRNA maximal expression in the evening or midnight, while CLOCK mRNA expression shows lack of rhythmicity (15, 17, 20). In the present study, the venous blood was collected once, in the morning between 06:00–10:00 hours, and we were, therefore, able to assess PER1, PER2, PER3, CRY1, and CRY2 mRNA expression at their approximate acrophase. At the same time, BMAL1 mRNA expression was in an antiphase to these genes. In addition, the highest transcript level in circulating blood leukocytes, measured in the morning, was observed for PER1, while the lowest was observed for PER3.
In the crude analysis of circadian gene expression in circulating leukocytes, we observed significant up-regulation of constitutive PER1 and PER3 mRNA expression in circulating leukocytes of nurses and midwives, who currently work rotating night shifts, compared to day workers. When we adjusted gene expression for significant confounding factors, the association between rotating shift work and circadian gene expression disappeared. However, the highest expression of PER1 mRNA was observed for women currently working shifts who had worked rotating night shift schedule >15 years compared with day workers who had previously worked rotating shifts >15 years. Moreover, the results of our study suggested that up-regulation of PER1 mRNA expression may be associated with the lifetime duration of night shift work among nurses and midwives who currently work rotating night shifts. These findings could result from a cumulative effect of night shift work on PER1 gene expression, associated with phase delay or advance of maximal circadian gene expression or direct effect of artificial light at night exposure. It is widely known that resetting of the peripheral clocks can occur through regulation of the expression of clock genes caused by internal or external cues. Generally, light exposure in the morning advances and light exposure in the evening delays circadian systems in organisms, which may cause serious effects due to irregular and disturbed wake–sleep cycle of humans (26). It was found that shift work caused a delay of the clock gene’s acrophase in human peripheral tissues. Results of a study on circadian gene mRNA expression (NR1D1, NR1D2, PER2, and PER3) in head hair follicle cells of rotating shift workers showed circadian gene expression phase delay after the second week of late shift work (27). Under short-term acute sleep deprivation (40 hours) and light conditions, PER2 mRNA expression in PBMC presented delayed phase shift and loss of circadian rhythmicity, while BMAL1 mRNA expression did not show any changes (28). James et al (22) found that a shifted sleep–wake schedule with light at night exposure significantly affects circadian expression of PER1 and PER2 peripheral circadian oscillators in PBMC and centrally-driven melatonin secretion. Blue light reduced BMAL1 mRNA expression in PBMC of jaundiced neonates under light therapy with covered eyes (29), but short wavelength light at 460 nm (blue light) induced PER2 expression in oral mucosa during a 2-hour exposure of human subjects in the evening (30).
Circadian gene expression may be also regulated by both photic and circadian inducers because it has been widely observed that other clues, such as dietary and feeding habits (31), alcohol use (10), depression (32), age (33), chronotype (18), may also influence peripheral circadian rhythm measured in peripheral leukocytes. In our cross-sectional study, no association has been observed between BMI, fasting status, alcohol drinking, and circadian gene expression.
Interestingly, we observed that PER1 and PER3 transcripts level was significantly down-regulated in the late versus early hours of the morning. The analysis of time of blood collection in the morning indicated that blood samples from rotating shift women were collected at 07:15 (SD 0.37) hours and 08:30 (SD 0.50) hours for day workers. This 1.15-hour difference in blood sampling of women under two work schedules may result from observed differences in PER1 and PER3 gene expression in the crude analysis. Based on rhythmic expression of circadian genes in various peripheral tissues, we might have observed increasing expression or acrophase of these two genes in the leukocytes of rotating shift women at 07:15 hours, while – among day workers – measured expression could have been attributable to decreasing phase at 08:30 hours. Due to highly significant collinearity of night shift work status and time of blood collection, we were unable to distinguish whether the effect we saw was attributed to the former or the latter. In comparison, the results from the only study on circadian genes mRNA expression analyzed in peripheral blood cells (collected at approximately 09:00 hours) of 25 Japanese shift workers and 45 participants who kept regular hours showed that the PER3 transcripts level were higher among shift working women compared to day workers (b-coefficient 0.31, P<0.05) (29).
In summary, in our study, we did not observe a significant association between circadian gene expression and rotating shift work; time of blood collection was found to be an important covariate in the analyses of PER1 and PER3 mRNA morning expression. Further studies with daily monitoring of circadian genes expression are needed to (i) confirm the impact of irregular working hours and light at night exposure on peripheral circadian clocks in humans and (ii) elucidate the molecular mechanisms of light-induced circadian gene alteration in human circulating leukocytes. Importantly, when analysing the peripheral clock in human studies, the hour of blood collection should be precisely specified.