Chronic obstructive pulmonary disease (COPD) is a common and complex disease with a genetic contribution (1). The most well-established risk polymorphisms are single-nucleotide polymorphisms (SNP) in the SERPINA gene causing alpha-1-antitrypsin deficiencies (2, 3). However, only a few percent of COPD cases can be explained by the presently identified genetic variants. Further genetic studies in COPD are therefore needed to identify new genetic variants in order to establish new molecular targets for primary prevention, diagnosis, and treatment (4, 5).
Heme oxygenase 1 (HMOX1) is an anti-oxidant and anti-inflammatory heme-degrading enzyme and the gene is known as a cytoprotective stress response gene (6–8). A dinucleotide repeat (GT)n sequence in the 5’ of the promoter region of the HMOX1 gene (rs3074372) modulates gene expression. A long allele (many repeats) has been shown to reduce enzyme activity and associate with various diseases (8, 9). Several studies have previously discussed whether this polymorphism affects COPD and impairs lung function yet with divergent outcomes and conclusions (4).
The main risk factor for COPD is smoking, but several occupational exposures are causally related to COPD (10). Furthermore, a genome-wide interaction study of gene-by-occupational exposure has recently identified new genetic variants associated with the level of forced expiratory volume in the first second of expiration (FEV1), where seven single nucleotide polymorphisms interacted with one of the occupational exposures (11).
In a Danish population-based cohort of 45–84-year-old participants, we previously suggested occupational exposure to vapor, gas, dust, or fume (VGDF) is associated with COPD (12). In this paper, we hypothesise that the effect of a genetic variant may be different in occupationally exposed compared to smoking subjects. This is due to a lower inflammatory effect on lungs by occupational exposure than by individual smoking. Thus, the genetic effect may be more pronounced in unexposed subjects or less occupational exposed subjects.
The aim of this cross-sectional study was first to investigate the association of HMOX1 promoter polymorphism of (GT)n repeats with COPD and second to investigate the presence of a possible gene-by-environment interaction with VGDF and smoking.
Methods
Population
The study population is based on the Danish North Jutland COPD Prevention Study (NCPS), the primary aims of which were to estimate the prevalence of COPD in the Danish population aged 45–84 years and to determine the prevalence of cases without appropriate medical treatment (13). General practitioners (GP) from two former counties in northern Jutland enrolled participants in the period October 2004 – September 2006. Patients listed with participating GP were selected randomly from the Danish Civil Registration System based on the expected prevalence of COPD in sex-specific 10-year age groups. Thus, the sample a priori had an overweight of elderly participants and men. Data were gathered from self-administered questionnaires (on education, work, family history of lung disease, medical information, smoking and alcohol habits, asthma, and allergy) and medical examinations including spirometry and blood samples. The response rate in the study was 36% (13). We studied occupational VGDF exposure in this cohort and included 4717 participants (12). All participants of non-Scandinavian descent were identified through their names and excluded in the genetic part of the study (N=14).
Lung function and COPD definition
The GPs obtained measures of FEV1 and forced vital capacity (FVC) by spirometry. All healthcare staff performing spirometry participated in a one-day training module before entering the study, and all spirometers were calibrated twice every year. A bronchodilator was not administered to all participants; hence, pre-bronchodilator spirometry was used consistently throughout the study. The lower limit of normal (LLN) approach was used to define COPD (14) in a screening setting based on the FEV1/FVC 2.5th centile (z-score = -1.96) (15). COPD severity was assessed using a detailed grading system based on LLN according to FEV1. To distinguish subjects with at least moderate airway obstruction, we added the recommended criterion of LLN for FEV1 of a z-score = -2.0 (rounded 2.5th centile) (16). As reference population, we used the Global Lung Function 2012 Equations (15) in the “GLI-2012 Desktop Software for Large Data Sets version 1.3.4 build 3” (17).
Occupational exposure
The duration of occupational exposure (years in occupations with exposure) was used as a proxy for cumulative occupational exposure and assessed based on a self-administered questionnaire of past and present occupations. The lifetime occupational VGDF exposure was divided into: organic dust, inorganic dust, vapor, and gas/fume. To improve the validity of the self-reported exposure information, we restricted such information as to a priori expert-selected jobs with known occupational exposures to VGDF using the Danish adapted version of the International Standard Classification of Occupations, revision 1988 (DISCO-88) (18) (supplementary material www.sjweh.fi/show_abstract.php?abstract_id=3846, table S1). The expert-selected jobs with occupational exposure included 72 of 372 DISCO-88 codes. VGDF exposure was originally assessed in four levels: never, low (<5 years), medium (5–14 years), and high (≥15 years) occupational lifetime exposure. VGDF was used as a dichotomized never versus ever (low/medium/high) exposure variable due to power issues when the genetic variant was included.
Genotyping
The repeat polymorphism in the promoter region in HMOX1 (rs3074372, GRCh38:22:35,381,066-35,394,213) was amplified by polymerase chain reaction (PCR) using a fluorescent-labelled sense primer (5’-AGAGCCTGCAGCTTCTCAGA-3’) and an unlabeled antisense primer (5’-ACAAAGTCTGGCCATAGGAC-3’) covering the (GT)n repeats (19–21). The PCR products were subsequently genotyped according to size using an ABI 3730XL DNA Analyzer (Thermo Fischer Scientific, Waltham, MA, USA) using GeneMarker® software (SoftGenetics, State College, PA, USA). The output of each allele GT repeats was blinded and calculated in respect to an internal size standard where 128 base pairs equals 30 GT repeats. Of the blood samples, 31 were missing, 48 were insufficient (eg, serum) and 201 failed in the HMOX1 analysis, which resulted in 4423 participants with a valid HMOX1 genotype. The cut-off of the GT repeats was defined as short (S): ≤26; medium (M): 27−32; and long (L): ≥33 GT repeats (21–23). The individual S/M/L genotype was grouped according to an L-dominant genetic model as HMOX1 L- (S/S, S/M, M/M) and HMOX1 L+ (S/L, M/L, L/L) groups.
Smoking
Cumulated smoking estimated as smoked pack-years was categorized into three levels, namely, <10, 10–20, and >20 pack-years. Different types of smoking were converted into number of smoked cigarettes as follows: one cheroot=three cigarettes; one cigar=four cigarettes; one pipe bowl=three cigarettes; and a package of pipe tobacco (50 gram)=17 pipe bowls.
Statistics
Statistical analyses were conducted in Stata 14.2 (StataCorp LP, College Station, TX, USA). The significance level was set at 5%. The 95% confidence interval (CI) was calculated using a normal approximation. The chi-square test for categorical variables was used to assess differences between sub-groups of the study population. Status of Hardy-Weinberg Equilibrium (HWE) was performed among non-COPD participants according to their HMOX1 S/M/L genotype. Associations of the HMOX1 polymorphism with COPD were analyzed in univariate and mixed random effect logistic regression models (24) with GP practice as a random variable. Likelihood-ratio tests were performed for possible gene-by-environment interactions of occupational exposure and smoking, respectively. The odds ratio (OR) from the mixed regression model was adjusted for pack-year, sex, occupational exposure, and age as fixed effects. Additionally, analyses were presented in a separate table as recommended by Knol et al (25), including post-estimation of any additive interaction. We used five regression models to compare no COPD with each level of COPD severity. Sensitivity analyses were performed by restriction according to (i) never smokers; (ii) no prior self-reported asthma; (iii) different dichotomization of occupational VGDF exposure: never/low versus medium/high and never/low/medium versus high occupational exposure; and (iv) different HMOX1 L± cut-offs according to (GT)n promoter repeats (N=31, 32, 34 and 35). The arguments for these analyses were: (a) to emphasize the a priori assumption of a diverse effect of HMOX1 polymorphism between a large smoking exposure and a low occupational exposure, by excluding the major risk factor of COPD, (b) to avoid the predisposing effect of asthma to COPD in the analysis (26, 27), (c) to investigate the effect of different levels of occupational exposures in the model, and (d) to analyze the effect of different cut-offs of the HMOX1 repeat polymorphism due to no clear argument for the used HMOX1 cut-offs in the literature.
Ethics
The NCPS study was performed in accordance with the Helsinki Declaration and approved by the Danish Scientific Ethics Committee (VN2003/62) and the Danish Data Protection Agency (2007-41-1576). Written informed consent was obtained from all participants. The Danish Scientific Ethics Committee also approved blood usage in the genetic analyses (N-20130038).
Replication study
The analyses were replicated in the Danish RAV (risk factors for asthma among adults) cohort (28), which is based on the European Community Respiratory Health Survey (ECHRS) protocol (29). Briefly, the RAV cohort, which was designed primarily to identify risk factors for asthma, comprised 20–44-year-old subjects (N=1191) of whom 44% were males, 54% were never smokers, and 30% had ever self-reported asthma. Thirty-four COPD cases were present (3%). In addition, data on lung function, occupational history of the latest ten jobs and blood samples were available. Participants were excluded if they were born outside Scandinavia (N=23); 86 blood samples failed in the HMOX1 analysis. Hence, 1082 participants with a valid HMOX1 genotype were available for analysis.
Results
The study population is characterized in table 1 according to genotype and stratified by presence of the long allele (L± GT33 repeats). The population included 6% with COPD, 48% who had smoked ≥10 pack-years, and 46% with occupational VGDF exposure. Only the prevalence of asthma and COPD differed between the L- and the L+ genotypes. The (GT)n allelic distribution of the longest allele is shown in figure 1. An HMOX1 L+ genotype was present in 10.6%. HMOX1 L+ in non-COPD participants fulfilled the criteria of being in HWE, P=0.24. The crude association between COPD and a long (GT)n allele (HMOX1 L+) was OR 1.66 (95% CI 1.17-2.34) while the adjusted OR slightly increased OR 1.75 (95% CI 1.18–2.60) (table 2). Additionally increased OR of COPD was observed when having HMOX1 L+ and occupational VGDF exposure together in a gene-by-environment interaction, OR 2.38 (95% CI 1.04–5.46), versus HMOX1 S+M and no exposure. There seemed to be a positive additive interaction as well as a positive interaction on the multiplicative scale between HMOX1 L+ and VGDF exposure (table 3). As the estimated OR exceeded the expected OR, we additionally analyzed the data without any adjustments (supplementary table S2.A) and without the pack-year adjustment (supplementary table S2.B); these estimated OR were 2.75 (95% CI 1.30–5.80) and 2.62 (95% CI 1.15–5.95), respectively.
Table 1
Characteristics of the study participants N=4423 a stratified by HMOX1 ±long GT allele genotype. [COPD=chronic obstructive pulmonary disease; L- =no-polymorphism, <33 GT promoter repeats; L+ =(GT)n promoter repeat polymorphism, including ≥33 GT repeats; S=small ≤26 GT repeats; M=medium 27−32 GT repeats; L=long ≥33 GT repeats; VGDF=occupational vapor, gas, dust, or fumes]. Bold indicates statistically significant, P<0.05.
| HMOX1 GT33 promoter repeats | P value | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| COPD b | L- genotype | L+ genotype | ||||
|
|
|
|
||||
| N | N | % | N | % | ||
| HMOX1 genotype L- | 3956 | 89 | 467 | 11 | ||
| S/S | 37 | 534 | 14 | - | - | |
| S/M | 100 | 1875 | 47 | - | - | |
| M/M | 86 | 1547 | 39 | - | - | |
| HMOX1 genotypeb L+ | ||||||
| S/L | 16 | - | - | 172 | 37 | |
| M/L | 26 | - | - | 289 | 62 | |
| L/L | 0 | - | - | 6 | 1 | |
| Sex | ||||||
| Female | 73 | 1369 | 35 | 155 | 33 | |
| Male | 192 | 2587 | 65 | 312 | 67 | 0.5 |
| Age years | ||||||
| 45−54 | 29 | 802 | 20 | 96 | 21 | |
| 55−64 | 46 | 930 | 24 | 119 | 25 | |
| 65−74 | 122 | 1485 | 38 | 174 | 37 | |
| 75−84 | 68 | 739 | 19 | 78 | 17 | 0.7 |
| Smoked pack-years (N=4093) | ||||||
| 0<10 | 38 | 1785 | 49 | 197 | 46 | |
| 10−20 | 25 | 483 | 13 | 72 | 17 | |
| >20 | 177 | 1395 | 38 | 161 | 37 | 0.1 |
| VGDF exposure (N=4196) | ||||||
| No | 106 | 1951 | 52 | 236 | 54 | |
| Low | 19 | 317 | 8 | 32 | 7 | |
| Medium | 34 | 344 | 9 | 53 | 12 | |
| High | 90 | 1143 | 30 | 120 | 27 | 0.1 |
| Ever asthma (N=4365) | ||||||
| No | 148 | 3505 | 90 | 391 | 85 | |
| Yes | 110 | 401 | 10 | 68 | 15 | <0.01 |
| COPD b (N=4406) | ||||||
| No | 3718 | 94 | 423 | 91 | ||
| Yes | 265 | 223 | 6 | 42 | 9 | <0.01 |
Figure 1
The (GT)n allelic distribution of the longest allele among the 4423 valid HMOX1 analyses in the North Jutland COPD Prevention Study. The vertical dotted line divides the group into L- and L+ genotype according to an L-dominant genetic model. [S=small, ≤26 GT repeats; M=medium, 27−32 GT repeats; L=large, ≥33 GT repeats.]
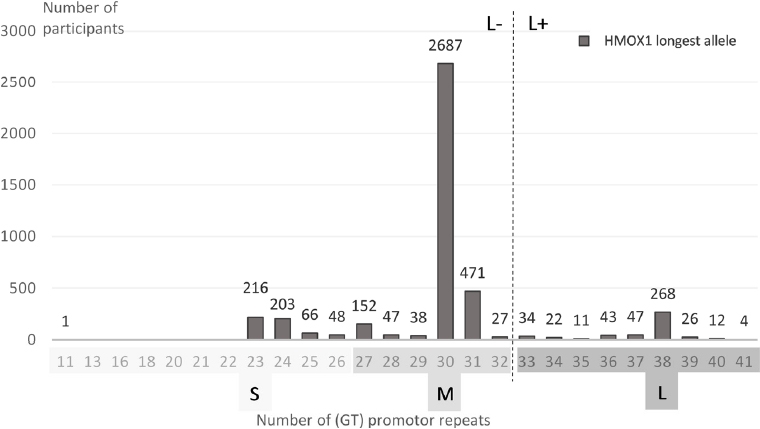
Table 2
Associations between chronic obstructive pulmonary disease (COPD) and Heme oxygenase 1 (HMOX1) polymorphism with and without the occupational vapor, gas, dust or fume (VGDF) exposure interaction in the North Jutland COPD Prevention Study (NCPS) and RAV (risk factors for asthma among adults cohort) studies. [CI=confidence interval; L- =no-polymorphism, <33 (GT) promoter repeats; L+ =(GT)n promoter repeat polymorphism, including ≥33 (GT) repeats; OR=odds ratio; NA=not available, too few observations; - =not included in the model]. Bold indicates statistically significant, P<0.05.
| NCPS (all, N=3871) | NCPS (never smokers, N=1385) | RAV replication study (N=986) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Adjusted OR a | Adjusted OR a+ interaction | Adjusted OR a | Adjusted OR a | |||||
|
|
|
|
|
|||||
| OR | 95% CI | OR | 95% CI | OR | 95% CI | OR | 95% CI | |
| HMOX1 | ||||||||
| L- | 1 | 1 | 1 | 1 | ||||
| L+ | 1.75 | 1.18–2.60 | 1.06 | 0.55–2.06 | 2.41 | 0.75–7.78 | 0.23 | 0.03–1.75 |
| Pack–years of smoking | ||||||||
| <10 | 1 | 1 | - | - | 1 | |||
| 10–20 | 2.69 | 1.57–4.60 | 2.70 | 1.58–4.61 | - | - | 2.59 | 0.95–7.04 |
| >20 | 7.64 | 5.13–11.39 | 7.67 | 5.15–11.42 | - | - | 3.60 | 1.29–10.00 |
| Sex | ||||||||
| Female | 1 | 1 | 1 | 1 | ||||
| Male | 0.54 | 0.37–0.78 | 0.55 | 0.38–0.79 | 0.32 | 0.12–0.88 | 1.68 | 0.73–3.86 |
| Age, RAV replication study | ||||||||
| 20–25 | - | - | - | - | - | - | 1 | |
| 26–30 | - | - | - | - | - | - | 1.02 | 0.26–3.99 |
| 31–35 | - | - | - | - | - | - | 0.50 | 0.11–2.32 |
| 36–40 | - | - | - | - | - | - | 0.89 | 0.24–3.29 |
| 41–47 | - | - | - | - | - | - | 0.86 | 0.23–3.29 |
| Age, NCPS study population | ||||||||
| 45–54 | 1 | 1 | 1 | |||||
| 55–64 | 1.29 | 0.76–2.17 | 1.32 | 0.78–2.23 | 1.16 | 0.25–5.46 | - | - |
| 65–74 | 2.26 | 1.41–3.60 | 2.29 | 1.43–3.66 | 1.63 | 0.40–6.61 | - | - |
| 75–84 | 2.33 | 1.39–3.91 | 2.38 | 1.42–3.99 | 2.66 | 0.59–12.02 | - | - |
| VGDF | ||||||||
| Never | 1 | 1 | 1 | 1 | ||||
| Ever | 1.38 | 1.01–1.88 | 1.21 | 0.87–1.69 | 5.12 | 1.70–15.46 | 1.73 | 0.73–4.09 |
| VGDF and HMOX1 interaction | ||||||||
| Never/L- | - | - | 1 | NA | NA | |||
| Ever/L+ | - | - | 2.38 | 1.04–5.46 | NA | NA | NA | NA |
Table 3
Interaction between HMOX1 L± genotype and VGDF exposure on the risk of chronic obstructive pulmonary disease (COPD), adjusted for pack-years, sex, age and general practitioner practice (N=3871). [HMOX1=Heme oxygenase 1 gene; L-= no-polymorphism, <33 (GT) promoter repeats; L+= (GT)n promoter repeat polymorphism, including ≥33 (GT) repeats; OR=odds ratio; RERI= relative excess risk due to interaction; VGDF=vapour, gas, dust or fume occupational exposure]. Bold indicates statistically significant, P<0.05. Note: Measure of interaction on additive scale: RERIOR (95% CI) = 1.79 (0.18−3.40); P=0.03. Measure of interaction on multiplicative scale: ratio of ORs (95% CI) = 2.38 (1.04−5.46); P=0.04.
No significant interaction was seen between the HMOX1 L+ and smoking: HMOX1 and >20 pack-years OR 1.13 (95% CI 0.38–3.35), likelihood-ratio test P=0.98 (supplementary table S3).
Sensitivity analyses: (i) Restricted to never smokers, the crude association between COPD and HMOX1 L+ was increased. Excluding sex and age variables in the model among never smokers barely changed the association between COPD and HMOX1 L+, OR 2.39 (95% CI 0.76–7.55); however, the impact of VGDF exposure seemed to decrease, OR 3.35 (95% CI 1.24–9.05). (ii) Among non-asthmatics, the crude association between COPD and HMOX1 L+ was similar to that of all participants, OR 1.69 (95% CI 1.07–2.28). (iii) Changing the occupational exposure to never/low versus medium/high slightly increased the effect of the VGDF and HMOX1 interaction. Comparing with the high exposure the estimate decreased into insignificance. (iv) Different genetic HMOX1 L cut-offs were analyzed. The main cut-off was 33 GT repeats, as shown in figure 1 by the dotted line. Of all the different cut-off models, only the lowest cut-off L+31 was insignificant according to HMOX1. All other models were similar to the used model with 33 GT repeats (L+).
Analysis according to COPD severity and HMOX1 L+ revealed no associations or indication of a trend (data not shown).
Replication study
In the RAV cohort, 19% had occupational VGDF exposure. The median age was 35 (standard deviation 7.0) years and 3% (N=32) had COPD. The (GT)n allelic distribution in the RAV cohort across genotypes (L-: S/S 15%, S/M 47%, M/M 38% and L+: S/L 34%, M/L 63%, L/L 4%) was similar to the NCPS distribution shown in table 1. HMOX1 L+ in non-COPD participants fulfilled the criteria of being in HWE, P=0.74. The estimates are included in table 2, showing no replication of the genetic effect and no replication of the HMOX1−VGDF interaction associated with COPD. Later, we additionally used asthma as outcome in the analysis of the younger RAV cohort since asthma predisposes to COPD. We found an insignificant, positive association with HMOX1 L+, OR 1.15 (95% CI 0.76–1.74).
Discussion
We found an increased risk of COPD in subjects carrying ≥1 long GT allele in the HMOX1 alone and in a gene-environmental interaction with occupational VGDF exposure. The combined COPD effect exceeded the effect of occupational VGDF exposure alone, suggesting that the association of the long allele of HMOX1 with occupational VGDF exposure is stronger than the association of HMOX1 with smoking between which no significant interaction was observed. The results among never smokers tend to support the association between COPD and HMOX1 L+ (similar OR) and VGDF (increased OR) compared to the main model without gene-environmental interaction (table 2). As asthma predisposes to COPD development (26, 27), analyses performed among non-asthmatics might underestimate the true association. On the other hand, due to the same reason, inclusion of asthmatics might also have overestimated the association. Thus, the best epidemiological estimate probably lies somewhere between the estimates obtained with models including and excluding asthmatics. The insignificance occurred when changing the occupational exposure to never/low/medium versus high exposure could be explained by low power and/or a healthy worker effect (supplementary table S4).
We applied the commonly used cut-off of L+ ≥33 GT repeats for the HMOX1 promotor polymorphism. However, the model was slightly sensitive for change in HMOX1 L cut-offs between L32 and L35.
Comparable studies in European populations regarding COPD and the (GT)n repeats have been studied. Matokanovic et al (30) found no association between COPD and the length of the GT repeat polymorphism in a small (N=225) Croatian population. In that study, COPD was defined by the GOLD criteria, and the study used cut-offs in GT repeats (S≤25, M 26-31, L≥32) that were slightly different from ours. Although, Hersh et al (20) found no association between COPD and the previously used S, M, and L groups of GT repeats in HMOX1, they found associations with the two most common single-number of repeats (GT repeats 30 and 31) in two different study populations (case−control and pedigree family-based). These populations included <1000 participants each.
Guenegou et al (21) studied associations of GT repeats in HMOX1 and 8-year lung function decline in 749 French participants in the European Community Respiratory Health Survey. L+ allele carriers had a more rapid decline in FEV1/FVC, and this decline was significantly steeper among heavy smokers. Siedlinski et al (22) later replicated these results in a Dutch general population (N=1390). The FEV1 decline was significantly increased in the M/L+L/L genotype, and the decline remained significant in restricted analyses in ever and heavy smokers (22).
In table 3, VGDF+ exposure was stratified within the HMOX1 L± strata, and the estimated OR was 3.30 (95% CI 1.33−8.19) in the HMOX1 L+ exposure group. As smoked pack-years were equally distributed between HMOX1 L± (table 1) and the expected OR according to table 3 would be OR 2.90 (= 3.07/1.06) and lower than the estimated OR, we interpret this stratified estimate as an indication of some interaction between HMOX1 L+ and smoking. To validate this interpretation, we analyzed the data without any adjustments (supplementary table S2.A) and without the pack-year adjustment (supplementary table S2.B); the calculated estimated OR were 2.75 (95% CI 1.30−5.80) and 2.62 (95% CI 1.15–5.95), respectively. Since these OR were more in line with the expected values, the data suggest that HMOX1 L+ interacts to a smaller extent with smoking than with VGDF in a gene-by-environmental pathway with COPD, see supplementary table S3; interaction between HMOX1 and >20 pack-years OR 1.13 (95% CI 0.38–3.35). We interpret this as the intense smoking exposure is less modified by the presence of HMOX1 L+, than the low-dose environmental exposures, eg, occupational exposure. Both additive and multiplicative statistical interactions of VGDF and HMOX1 were present (table 3). The additive estimate of interaction suggests the presence of a mechanistic interaction as a sufficient cause interaction, equally the relative excess risk due to interaction (RERI)>1. Although, RERI is not significant as the lower 95% CI are <1. Nevertheless, the biological role of HMOX1 appears rather well documented (31, 32), suggesting that our finding does represent a biological effect.
Two studies combined the promoter GT repeats in HMOX1 and lung function with other exposures of ozone (33) and PM10 (particulate matter <10 µm) (23). These studies point in the same direction as our study despite different L cut-offs, exposures and outcomes. Thus indicative of, a HMOX1 promoter polymorphism association with exogenous exposure.
Strengths and limitations
The strengths of the present study are its size, the expert-validated occupational exposures, and that genotyping was validated in a blinded setting. Ethnic variability was restricted although non-differential misclassification might occur. Another limitation is that we did not include other genetic risk factors that might affect the association with the VGDF exposure and HMOX1 interaction.
Of the samples in the HMOX1 analysis, 201 (NCPS) and 86 (RAV) failed, however, we deem the analytical success rate (95%) to be acceptable. The failed HMOX1 data not included in the analysis were equally distributed among plates. Among the study participants, there were more young women and fewer participants in the oldest group than among non-responders. This might introduce an age-dependent healthier study population, which would tend to underestimate the associations. The use of a spirometrically defined outcome of COPD implies that some COPD patients with compliance difficulties might have been excluded from the analysis, resulting in biased associations. Possible information bias of occupational VGDF exposure was reduced by using independent, specialist-assessed exposure based on job titles. The external study validity is considered good in a Western world setting with a similar occupational distribution and a Northern European genetic structure.
Replication
Although the distribution of HMOX1 genotypes was similar in RAV and NCPS, no associations were seen in the RAV cohort. We attribute this to the smaller study size and lower VGDF exposure in RAV and the short time period for development of COPD in this cohort. A priori, we assumed that the HMOX1 repeat polymorphism would be associated with COPD and not interact with occupational exposure. However, in the younger RAV cohort with less occupational exposure, HMOX1 L+ was inversely associated with COPD, and our data regrettably lacked power to analyze for the combined exposure. Although insignificant, the finding according to asthma might suggest that in this young cohort, an association could be relevant between HMOX1 L+ and obstructive lung disease.
To our knowledge, this is the largest population-based study analyzing promoter repeats in HMOX1. However, our findings would benefit from other replication studies with more VGDF exposure in an older cohort than the present study.
The intension of social threshold values as a tool for primary prevention is that workers doing their job will not be at increased risk of disease from occupational exposure, not even if they are genetically susceptible. The data may suggest that the actual threshold values ought to be lowered to meet the intension of the current labor market regulations. Moreover, the genetic marker may inform secondary prevention measures in workers with lung symptoms exposed to VGDF. Subjects with long GT alleles in HMOX1 may be more susceptible and would benefit from shifting to other work tasks with less VGDF exposure.
Concluding remarks
The present data support an association between subjects with a long GT allele in HMOX1 and COPD. A long GT allele in HMOX1 seems furthermore to interact with occupational VGDF exposure in a gene-by-occupational setting. We propose that the positive association with a long GT allele in HMOX1 may be a new genetic contributor to the pathogenesis of COPD. Additionally, the HMOX1 interaction with occupational VGDF exposure provide an argument for VGDF exposure level regulation in respect of primary and secondary prevention.



